ABSTRACT
Influenza A virus is a major causative agent of respiratory diseases in humans. It causes significant morbidity, mortality and economic losses each year worldwide with about 3-5 million clinical infections per annum. Its ability to mutate rapidly leads to seasonal epidemics and with its high frequency of genetic reassortment it causes pandemics. Conventional methods of studying viral pathogenesis do not allow for monitoring viral spread in real time during an infection.
Whole body bioluminescent imaging of infected animals will overcome many of the problems associated with the current methods. In this work the wild type Influenza A/WSN/33 was engineered to carry a luciferase reporter gene in segment 1, based on a well established reverse genetics system for Influenza A viruses. The novel reporter WSN virus will enable less expensive, non-invasive in vivo imaging of viral replication and better evaluation of novel therapeutics.
BACKGROUND
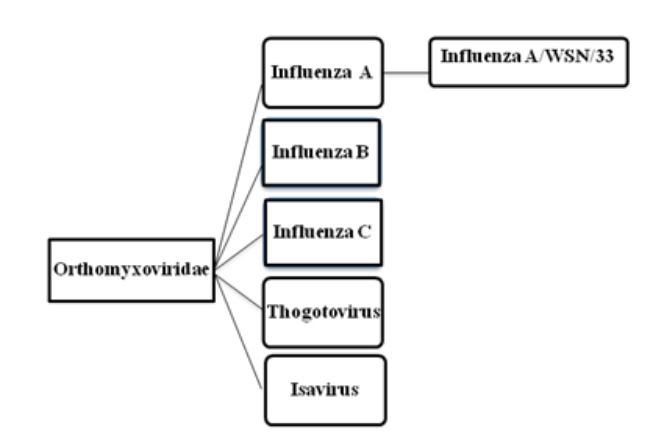
Figure 2.1: Taxonomy of Orthomyxoviruses
Each of the eight segments of Influenza A virus codes for at least one protein. When more than one protein is encoded by a single RNA segment, the virus hijacks the host-cell splicing machinery. The thirteen known viral proteins are summarized in table 2.1.
DESIGNING THE VIRUS
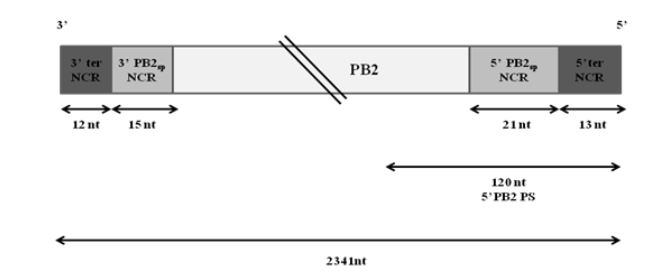
Figure 3.1: Schematic representation of wt PB2 vRNA of WSN virus
Extensive studies have been done to map the packaging signals on segment 1. The following highly conserved nucleotides of segment 1 have been shown to be crucial for proper packaging of WSN PB2 vRNA-nucleotides 2218 to 2220, nucleotides 2221 to 2224 and nucleotides 2296 to 2298. Fig. 3.1 illustrates the non-coding regions and the position of the 5′ packaging signal on PB2 vRNA.
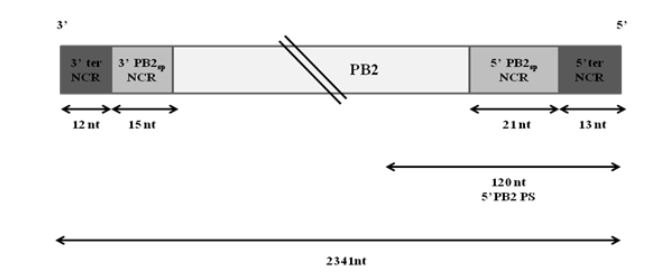
Figure 3.2: Schematic representation of the bi-directional plasmid pHW-PB2
The amplicons from these three PCR reactions had overhangs that could act as separate primers in the overlapping PCR reaction, PCR #4 to produce the whole of insert (ii) as shown in Fig. 3.2.
MATERIALS AND METHODS
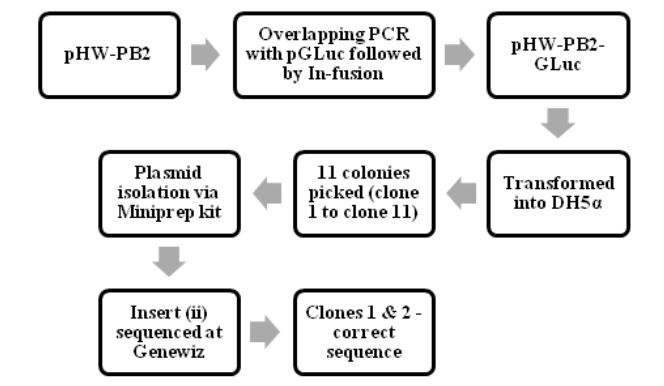
Figure 4.1: The process of making pHW-PB2-GLuc and cloning it into E.coli cells
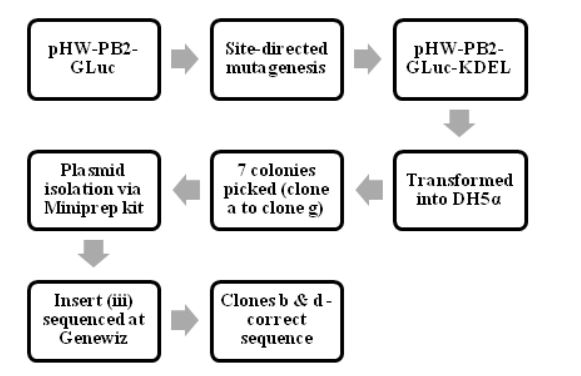
Figure 4.2: The process of making pHW-PB2-GLuc-KDEL and cloning it into E.coli cells
The various steps in making and characterizing the two recombinant WSN viruses are summarized in Fig. 4.1, Fig. 4.2 and Fig 4.3. The process of making pHW-PB2-GLuc and cloning it into E.coli cells. The wt WSN plasmid pHW-PB2 was used to make the mutant plasmid pHW-PB2-GLuc as described in section 3.3.2. Competent bacterial cells (DH5 α E.coli cells) were transformed with this plasmid.
RESULTS AND DISCUSSION
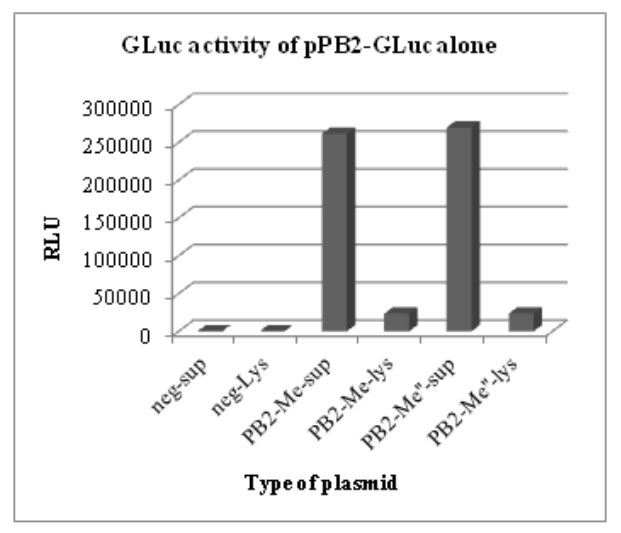
Figure 5.1: GLuc activity of pHW-PB2-GLuc alone. The supernatant and cell lysate from negative control
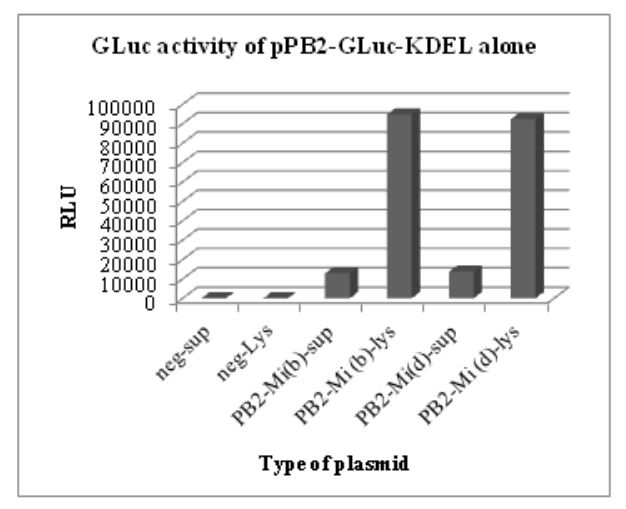
Figure 5.2: GLuc activity of pHW-PB2-GLuc-KDEL alone. The cell lysate and supernatant from negative control
The results of these luciferase assays are shown in Figs. 5.1 and 5.2. To test for the luciferase activity of the recombinant plasmids in the presence of the seven other plasmids (wild type) in the WSN reverse genetics system, different wells in a 6-well plate with a monolayer of 293T cells were co-transfected with the wild type plasmids for segment 2 to segment 8 in the WSN reverse genetics system.
CONCLUSION
Influenza A virus causes significant loss to humans each year due to its high mutability and frequent genetic reassortment capability. There is a continuing need to better understand the pathogenesis of the virus. Novel tools designed in this direction are invaluable. Whole-body bioluminescent imaging is one such tool that is becoming increasingly popular in the study of different diseases. In this work, the wild type Influenza A/WSN/33(H1N1) was successfully engineered for use in in vivo bioluminescent imaging.
The strategy employed to engineer the wild type lab strain of WSN/33 virus resulted in two recombinant WSN viruses with an integrated gene encoding the Gaussia luciferase enzyme. Both the viruses were shown to be replication-competent in vitro and to exhibit strong bioluminescent properties. The first mutant virus (recWSN-PB2-GLuc virus) can be used for high-throughput viral quantification, drug screening, anti-body characterization and other studies in vitro for better understanding the life cycle of Influenza A virus.
Source: University of California
Author: Chetan Raj Lingaraju